The technological advances in this techno-savy age are gradually converting this globe into a Virtual 3D World, where people prefer to view everything in a tertiary structure; right from playing a 3D video game to a 3D painting to designing a 3D protein structure. In medical field, 3D printing has many implications such as designing customized prosthetics, tissue and organ fabrication,etc.( Medical 3D printing )
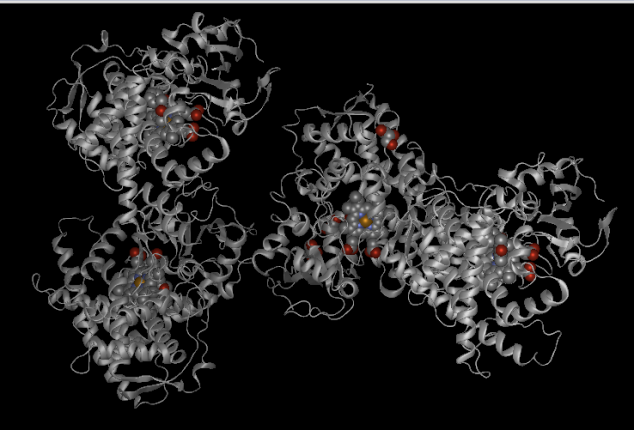
Visualizing a 3D structure of a gene or protein can be very useful from a bioinformatics perspective. It can be helpful for rational drug design, viewing sites associated with disease causing variants and also to see the proximity of amino acids that are distant in the primary sequence.As discussed in my previous post, the CYP2C19 gene is a monomer and a member of Cytochrome P450 family and is responsible for the metabolism of drugs such as anti-convulsant mephenytoin, anti-ulcer omeprazole and anti-malarial drug proguanil. An OMIM allele for CYP2C19 gene is.0003 ( p.TRP212TER (rs4986893) [dbSNP:rs4986893] ). A G to A mutation results in substitution of Tryptophan W at position 212 of protein sequence to a termination codon(X).This mutation leads to poor metabolism of proguanil and other drugs in individuals who are carriers for this variant.Using Protean 3D feature of DNA Star software we can visualize the 3D structure of Human CYP2C19 gene as follows:
In the above figure we can see a mixed backbone structure of gene which includes both ribbon and sphere design. From the style menu we can select different styles for the protein structure such as tube, balls and spheres, sticks, etc to customize it as per our visualization needs. Also below the 3D structure , there is the translated sequence of protein available along with the KSD secondary structure. We can highlight a portion of the sequence and the same gets highlighted in the tertiary structure, which helps us to gain an insight into the positioning of the amino acids i.e whether they are located on the surface or in the turn region or inside the helix.
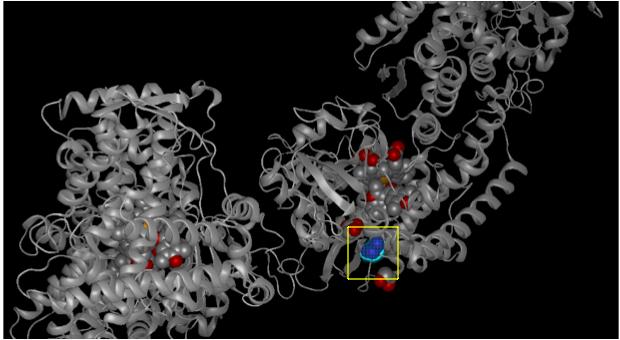
When we highlight our variant location for allele 0.0003, i.e amino acid Tryptophan at position 212, it gets reflected in the 3D structure as shown in figure below:
From this view, it can be seen that the variant position is located on the interior region (beta sheet ) of the helix. We can also see the variant location in different views and colors using the style menu :
Protean 3D allows us to use our creativity at its best and create different 3D view structures using various colors and design options available:
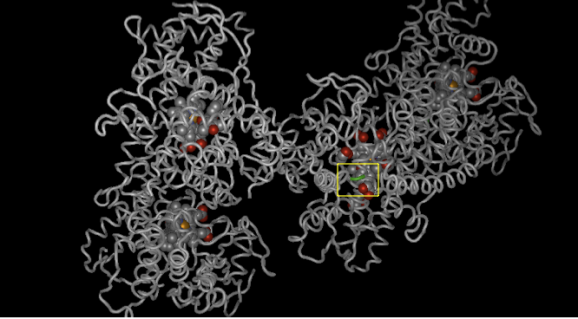
Tube temperature structure view of Human CYP2C19 gene with highlighted allele position in green. The spheres seen in the above image represent the ligands and water in the structure.
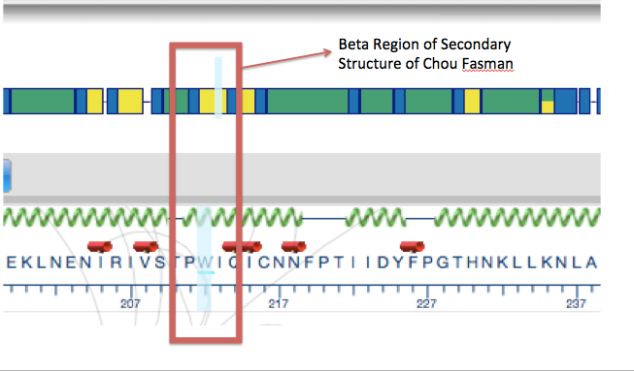
In order to check the validity of protein structure prediction algorithm, we compared the tertiary structure of Human CYP2C19 gene to the secondary structure prediction Chou Fasman algorithm in the analysis view of Protean 3D:
The above figure indicates that our 3D structure prediction is valid, as even the Chou Fasman secondary structure prediction algorithm shows that the variant site which is highlighted in light blue is located in the yellow color beta region of the helix.
When we looked at the Hydrophilicity plots generated by Kyte-doolittle and Hopp Woods algorithm, it further confirmed that the 3D structure prediction algorithm does tally with the secondary structure:
Both the hydrophilicity plots show that the highlighted variant site falls in the orange region which is representative of hydrophobicity. Thus the allele position is located in a hydrophobic region which is located usually in the lipid bilayer in the interior of the helix.
The Emini algorithm predicts the surface probability for a highlighted region and for position 212, which is the variant site in OMIM allele .0003 of the Human CYP2C19 gene, the surface region indicated is line as shown in the image below:
The violet blocks represent the surface whereas the lines are for the interior beta sheet regions.The light blue highlighted variant site falls in the line area, confirming our results with other predictive algorithms.
Thus, Protean 3D not only helps us to view the tertiary structure of a human gene or protein , but also through its analysis feature it allows us to compare the validity of different predictive algorithms in order to confirm our results and to give accurate structure prediction using different style options.
click on the following link for more information on types of protein structure and their implications in clinical filed :